
Sightings of Homo sapiens cognatus, or Homo sylvestris, are well-recorded, with a particular prevalence in North America. Bigfoot, otherwise known as the Sasquatch, is one of the most widely-reported cryptids in the world; it is the subject of documentaries, film, music, and countless books.
The Bigfoot Field Research Organisation has compiled a detailed database of Bigfoot sightings going back to the 1920s. Each sighting is dated, located to the nearest town or road, and contains a full description of the sighting. In many cases, sightings are accompanied by a follow-up report from the organisation itself.
As previously with UFO sightings and paranormal manifestations, our first step is to retrieve the data and parse it for analysis. Thankfully, the bfro.net dataset is relatively well-formatted; reports are broken down by region, with each report following a mainly standard format.
As before, we rely on the rvest package in R to explore and scrape the website. In this case, the key elements were to retrieve each state’s set of reports from the top level page, and retrieve the link for each report. Conveniently, these are in a standard format; the website also allows a printer-friendly mode that greatly simplifies scraping.
The scraping code is given here:
With each page retrieved, we step through and parse each report. Again, each page is fairly well-formatted, and uses a standard set of tags for date, location, and similar. The report parsing code is given here:
With each report parsed into a form suitable for analysis, the final step in scraping the site is to geolocate the reports. As in previous posts, we rely on Google’s geolocation API. For each report, we extract an appropriate address and parse it into a set of latitude and longitude coordinates. For the purposes of this initial scrape we restrict ourselves to North America, which compromises a large majority of reports on bfro.net. Geolocation code is included below. (Note that a Google Geolocation API key is required for this code to run.)
With geolocated data in hand, we can now venture into the wilds. In which areas of North America are Sasquatch most commonly seen to roam? The plot below shows the overall density of Bigfoot sightings, with individual reports marked.
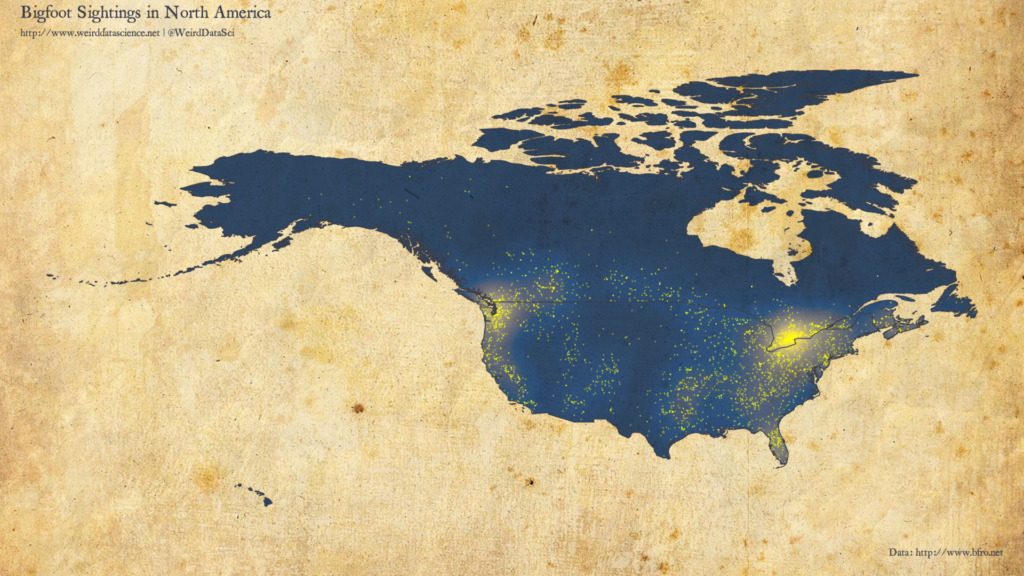
There are particular clusters on the Great Lakes, particularly in Southern Ontario; as well as in the Pacific Northwest. Smaller notable clusters exist in Florida, centered around Orlando. As with most report-based datasets, sightings are skewed towards areas of high population density.
The obvious first question to ask of such data is which, if any, environmental features correlate with these sightings. Other analyses of Bigfoot sightings, such as the seminal work of Lozier et al.1, have suggested that forested regions are natural habitats for Sasquatch.
To answer this, we combine the underlying mapping data and Bigfoot sightings, with bioclimatic data taken from the Global Land Cover Facility. Amongst other datasets, this provides us with an accurate, high-resolution land cover raster map, detailing vegetation for each 5-arcminute cell in the country — approximately one cell per 10km² .
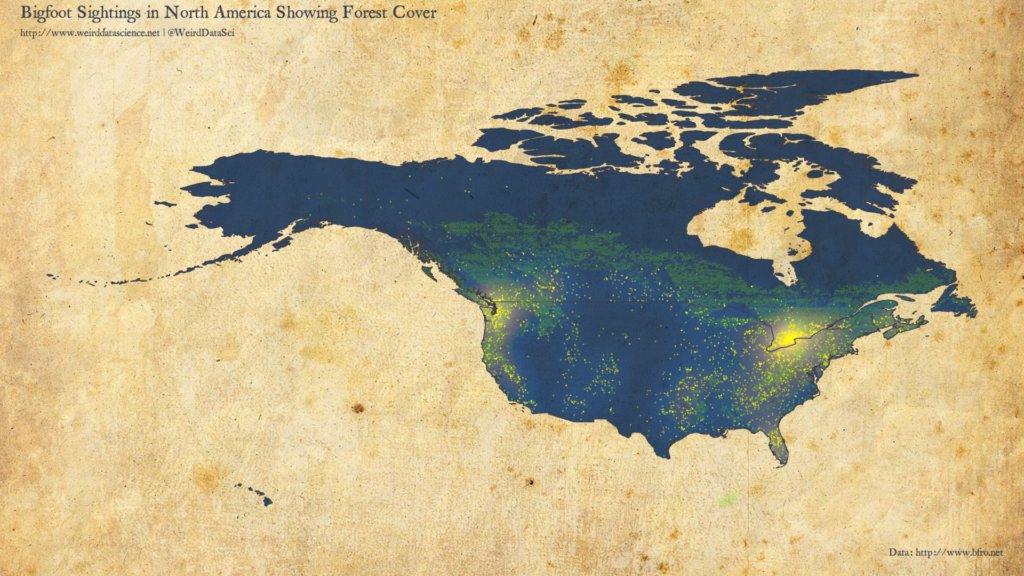
There are a range of bioclimatic variables in this dataset. The diagram below overlays all areas that are some form of forest onto the previous density plot.
The code for producing both of the above plots is given here:
From this initial plot we can see that, whilst tree cover is certainly not a bad predictor of Bigfoot sightings, it is far from a definitive correlation. The largest cluster, around the US-Canada border near Toronto, is principally lakes; whilst the secondary cluster in Florida is neither significantly forested or even close to the Everglades, which might have been expected. From the other perspective, there are significant forested areas for which sightings are reasonably rare.
The mystery of Bigfoot’s natural habitat and preferences is, therefore, very much unanswered from our initial analysis. With a broad range of variables still to explore — climate, altitude, food sources — future posts will attempt to determine what conditions lend themselves to strange survivals of pre-human primate activity. Perhaps changing conditions have pushed our far-distant cousins to previously unsuspected regions.
Until then, we keep following these trails into data unknown.
References
- Lozier, J. D., Aniello, P. and Hickerson, M. J. (2009), Predicting the distribution of Sasquatch in western North America: anything goes with ecological niche modelling. Journal of Biogeography, 36: 1623-1627. doi:10.1111/j.1365-2699.2009.02152.x (http://onlinelibrary.wiley.com/doi/abs/10.1111/j.1365-2699.2009.02152.x)

Be the first to comment
This site uses User Verification plugin to reduce spam. See how your comment data is processed.